mat1 <- matrix(1:16,nrow=4)
mat1 [,1] [,2] [,3] [,4]
[1,] 1 5 9 13
[2,] 2 6 10 14
[3,] 3 7 11 15
[4,] 4 8 12 16June 1, 2024
June 4, 2025
A matrix is a rectangular collection of the same data type (see Figure 8.1). It can be viewed as a collection of column vectors all of the same length and the same type (i.e. numeric, character or logical) OR a collection of row vectors, again all of the same type and length. A data.frame is also a rectangular array. All of the columns must be the same length, but they may be of different types. The rows and columns of a matrix or data frame can be given names. However these are implemented differently in R; many operations will work for one but not both, often a source of confusion.
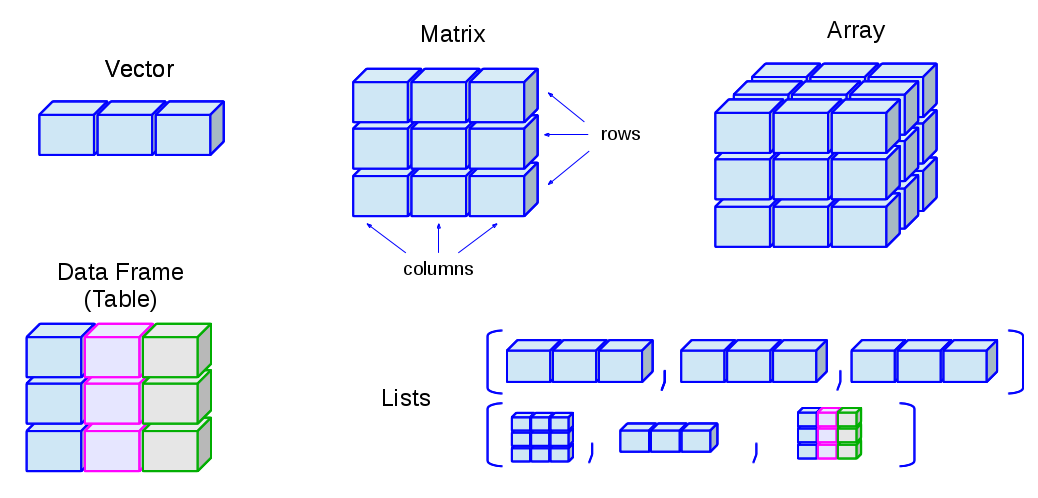
There are many ways to create a matrix in R. One of the simplest is to use the matrix() function. In the code below, we’ll create a matrix from a vector from 1:16.
mat1 <- matrix(1:16,nrow=4)
mat1 [,1] [,2] [,3] [,4]
[1,] 1 5 9 13
[2,] 2 6 10 14
[3,] 3 7 11 15
[4,] 4 8 12 16The same is possible, but specifying that the matrix be “filled” by row.
mat1 <- matrix(1:16,nrow=4,byrow = TRUE)
mat1 [,1] [,2] [,3] [,4]
[1,] 1 2 3 4
[2,] 5 6 7 8
[3,] 9 10 11 12
[4,] 13 14 15 16Notice the subtle difference in the order that the numbers go into the matrix.
We can also build a matrix from parts by “binding” vectors together:
x <- 1:10
y <- rnorm(10)Each of the vectors above is of length 10 and both are “numeric”, so we can make them into a matrix. Using rbind binds rows (r) into a matrix.
mat <- rbind(x,y)
mat [,1] [,2] [,3] [,4] [,5] [,6] [,7]
x 1.000000 2.00000000 3.000000 4.0000000 5.0000000 6.000000 7.000000
y -1.151193 -0.09401175 1.016198 -0.1788507 -0.9842443 -1.214426 -1.089794
[,8] [,9] [,10]
x 8.00000 9.0000000 10.0000000
y -2.23542 -0.4612952 0.4178649The alternative to rbind is cbind that binds columns (c) together.
mat <- cbind(x,y)
mat x y
[1,] 1 -1.15119280
[2,] 2 -0.09401175
[3,] 3 1.01619820
[4,] 4 -0.17885066
[5,] 5 -0.98424426
[6,] 6 -1.21442596
[7,] 7 -1.08979410
[8,] 8 -2.23542017
[9,] 9 -0.46129523
[10,] 10 0.41786492Inspecting the names associated with rows and columns is often useful, particularly if the names have human meaning.
We can also change the names of the matrix by assigning valid names to the columns or rows.
[1] "apples" "oranges"mat apples oranges
[1,] 1 -1.15119280
[2,] 2 -0.09401175
[3,] 3 1.01619820
[4,] 4 -0.17885066
[5,] 5 -0.98424426
[6,] 6 -1.21442596
[7,] 7 -1.08979410
[8,] 8 -2.23542017
[9,] 9 -0.46129523
[10,] 10 0.41786492Matrices have dimensions.
Indexing for matrices works as for vectors except that we now need to include both the row and column (in that order). We can access elements of a matrix using the square bracket [ indexing method. Elements can be accessed as var[r, c]. Here, r and c are vectors describing the elements of the matrix to select.
The indices in R start with one, meaning that the first element of a vector or the first row/column of a matrix is indexed as one.
This is different from some other programming languages, such as Python, which use zero-based indexing, meaning that the first element of a vector or the first row/column of a matrix is indexed as zero.
It is important to be aware of this difference when working with data in R, especially if you are coming from a programming background that uses zero-based indexing. Using the wrong index can lead to unexpected results or errors in your code.
# The 2nd element of the 1st row of mat
mat[1,2] oranges
-1.151193 # The first ROW of mat
mat[1,] apples oranges
1.000000 -1.151193 # The first COLUMN of mat
mat[,1] [1] 1 2 3 4 5 6 7 8 9 10# and all elements of mat that are > 4; note no comma
mat[mat>4][1] 5 6 7 8 9 10## [1] 5 6 7 8 9 10Note that in the last case, there is no “,”, so R treats the matrix as a long vector (length=20). This is convenient, sometimes, but it can also be a source of error, as some code may “work” but be doing something unexpected.
We can also use indexing to exclude a row or column by prefixing the selection with a - sign.
mat[,-1] # remove first column [1] -1.15119280 -0.09401175 1.01619820 -0.17885066 -0.98424426 -1.21442596
[7] -1.08979410 -2.23542017 -0.46129523 0.41786492mat[-c(1:5),] # remove first five rows apples oranges
[1,] 6 -1.2144260
[2,] 7 -1.0897941
[3,] 8 -2.2354202
[4,] 9 -0.4612952
[5,] 10 0.4178649We can create a matrix filled with random values drawn from a normal distribution for our work below.
V1 V2
Min. :-1.8330 Min. :-2.0497
1st Qu.:-0.7606 1st Qu.:-0.4389
Median : 0.7864 Median : 0.6033
Mean : 0.2856 Mean : 0.6738
3rd Qu.: 0.9019 3rd Qu.: 1.7985
Max. : 2.3502 Max. : 2.9927 Multiplication and division works similarly to vectors. When multiplying by a vector, for example, the values of the vector are reused. In the simplest case, let’s multiply the matrix by a constant (vector of length 1).
# multiply all values in the matrix by 20
m2 = m*20
summary(m2) V1 V2
Min. :-36.661 Min. :-40.994
1st Qu.:-15.211 1st Qu.: -8.779
Median : 15.729 Median : 12.065
Mean : 5.711 Mean : 13.475
3rd Qu.: 18.038 3rd Qu.: 35.969
Max. : 47.004 Max. : 59.854 By combining subsetting with assignment, we can make changes to just part of a matrix.
# and add 100 to the first column of m
m2[,1] = m2[,1] + 100
# summarize m
summary(m2) V1 V2
Min. : 63.34 Min. :-40.994
1st Qu.: 84.79 1st Qu.: -8.779
Median :115.73 Median : 12.065
Mean :105.71 Mean : 13.475
3rd Qu.:118.04 3rd Qu.: 35.969
Max. :147.00 Max. : 59.854 A somewhat common transformation for a matrix is to transpose which changes rows to columns. One might need to do this if an assay output from a lab machine puts samples in rows and genes in columns, for example, while in Bioconductor/R, we often want the samples in columns and the genes in rows.
t(m2) [,1] [,2] [,3] [,4] [,5] [,6] [,7]
[1,] 116.89722 147.004355 116.19088 99.50672 121.81061 63.33906 118.417923
[2,] 59.85408 -9.877637 12.99534 -40.99377 51.33027 -11.52863 -5.482144
[,8] [,9] [,10]
[1,] 78.79549 115.26654 79.88263
[2,] 11.13558 29.04172 38.27873Again, we just need a matrix to play with. We’ll use rnorm again, but with a slight twist.
Since these data are from a normal distribution, we can look at a row (or column) to see what the mean and standard deviation are.
mean(m3[,1])[1] 6.709692sd(m3[,1])[1] 1.425251# or a row
mean(m3[1,])[1] 4.86667sd(m3[1,])[1] 2.102434There are some useful convenience functions for computing means and sums of data in all of the columns and rows of matrices.
colMeans(m3) [1] 6.709692 4.968957 5.961377 4.572742 5.293570 4.130627 4.195079 4.475901
[9] 5.076318 5.360578rowMeans(m3) [1] 4.866670 4.100055 4.814678 5.800869 5.813489 5.485770 4.889247 5.444376
[9] 4.603745 4.925941rowSums(m3) [1] 48.66670 41.00055 48.14678 58.00869 58.13489 54.85770 48.89247 54.44376
[9] 46.03745 49.25941colSums(m3) [1] 67.09692 49.68957 59.61377 45.72742 52.93570 41.30627 41.95079 44.75901
[9] 50.76318 53.60578We can look at the distribution of column means:
Min. 1st Qu. Median Mean 3rd Qu. Max.
4.131 4.500 5.023 5.074 5.344 6.710 Note that this is centered pretty closely around the selected mean of 5 above.
How about the standard deviation? There is not a colSd function, but it turns out that we can easily apply functions that take vectors as input, like sd and “apply” them across either the rows (the first dimension) or columns (the second) dimension.
Min. 1st Qu. Median Mean 3rd Qu. Max.
0.7965 1.8319 2.0329 1.8965 2.1867 2.3309 Again, take a look at the distribution which is centered quite close to the selected standard deviation when we created our matrix.
For this set of exercises, we are going to rely on a dataset that comes with R. It gives the number of sunspots per month from 1749-1983. The dataset comes as a ts or time series data type which I convert to a matrix using the following code.
Just run the code as is and focus on the rest of the exercises.
data(sunspots)
sunspot_mat <- matrix(as.vector(sunspots),ncol=12,byrow = TRUE)
colnames(sunspot_mat) <- as.character(1:12)
rownames(sunspot_mat) <- as.character(1749:1983)After the conversion above, what does sunspot_mat look like? Use functions to find the number of rows, the number of columns, the class, and some basic summary statistics.
Practice subsetting the matrix a bit by selecting:
sunspot_mat[1:10,]
sunspot_mat[,7]
sunspot_mat['1979',7]These next few exercises take advantage of the fact that calling a univariate statistical function (one that expects a vector) works for matrices by just making a vector of all the values in the matrix. What is the highest (max) number of sunspots recorded in these data?
max(sunspot_mat)And the minimum?
min(sunspot_mat)And the overall mean and median?
Use the hist() function to look at the distribution of all the monthly sunspot data.
hist(sunspot_mat)Read about the breaks argument to hist() to try to increase the number of breaks in the histogram to increase the resolution slightly. Adjust your hist() and breaks to your liking.
hist(sunspot_mat, breaks=40)Now, let’s move on to summarizing the data a bit to learn about the pattern of sunspots varies by month or by year. Examine the dataset again. What do the columns represent? And the rows?
# just a quick glimpse of the data will give us a sense
head(sunspot_mat)We’d like to look at the distribution of sunspots by month. How can we do that?
Assign the month summary above to a variable and summarize it to get a sense of the spread over months.
Play the same game for years to get the per-year mean?
Make a plot of the yearly means. Do you see a pattern?