25 Introduction to SummarizedExperiment
The SummarizedExperiment class is used to store rectangular matrices of experimental results, which are commonly produced by sequencing and microarray experiments. Each object stores observations of one or more samples, along with additional meta-data describing both the observations (features) and samples (phenotypes).
A key aspect of the SummarizedExperiment class is the coordination of the meta-data and assays when subsetting. For example, if you want to exclude a given sample you can do for both the meta-data and assay in one operation, which ensures the meta-data and observed data will remain in sync. Improperly accounting for meta and observational data has resulted in a number of incorrect results and retractions so this is a very desirable property.
SummarizedExperiment is in many ways similar to the historical ExpressionSet, the main distinction being that SummarizedExperiment is more flexible in it’s row information, allowing both GRanges based as well as those described by arbitrary DataFrames. This makes it ideally suited to a variety of experiments, particularly sequencing based experiments such as RNA-Seq and ChIp-Seq.
25.1 Anatomy of a SummarizedExperiment
The SummarizedExperiment package contains two classes: SummarizedExperiment and RangedSummarizedExperiment.
SummarizedExperiment is a matrix-like container where rows represent features of interest (e.g. genes, transcripts, exons, etc.) and columns represent samples. The objects contain one or more assays, each represented by a matrix-like object of numeric or other mode. The rows of a SummarizedExperiment object represent features of interest. Information about these features is stored in a DataFrame object, accessible using the function rowData(). Each row of the DataFrame provides information on the feature in the corresponding row of the SummarizedExperiment object. Columns of the DataFrame represent different attributes of the features of interest, e.g., gene or transcript IDs, etc.
RangedSummarizedExperiment is the “child”” of the SummarizedExperiment class which means that all the methods on SummarizedExperiment also work on a RangedSummarizedExperiment.
The fundamental difference between the two classes is that the rows of a RangedSummarizedExperiment object represent genomic ranges of interest instead of a DataFrame of features. The RangedSummarizedExperiment ranges are described by a GRanges or a GRangesList object, accessible using the rowRanges() function.
Figure 25.1 displays the class geometry and highlights the vertical (column) and horizontal (row) relationships.
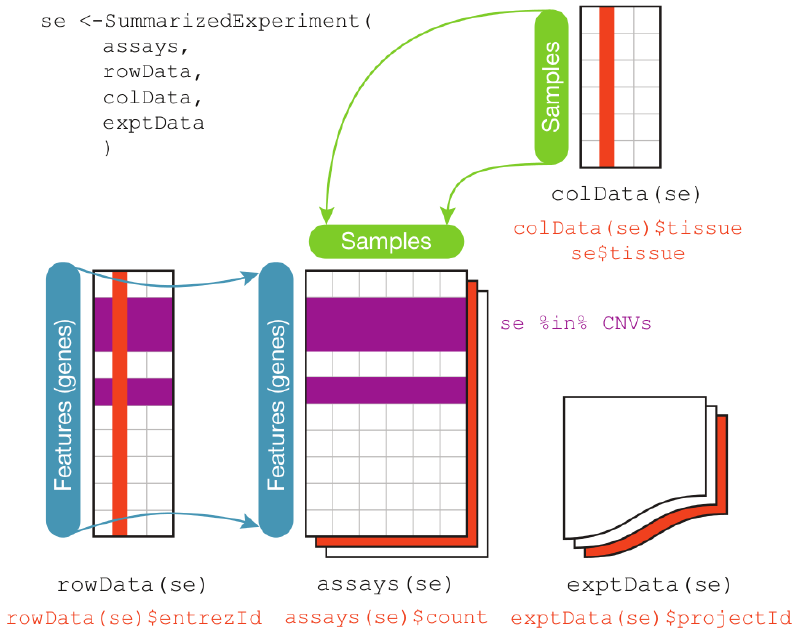
colData(), the rowData() and the assays(). The accessors for the various parts of a complete SummarizedExperiment object match the names.
25.1.1 Assays
The airway package contains an example dataset from an RNA-Seq experiment of read counts per gene for airway smooth muscles. These data are stored in a RangedSummarizedExperiment object which contains 8 different experimental and assays 64,102 gene transcripts.
Loading required package: airwaylibrary(SummarizedExperiment)
data(airway, package="airway")
se <- airway
seclass: RangedSummarizedExperiment
dim: 63677 8
metadata(1): ''
assays(1): counts
rownames(63677): ENSG00000000003 ENSG00000000005 ... ENSG00000273492
ENSG00000273493
rowData names(10): gene_id gene_name ... seq_coord_system symbol
colnames(8): SRR1039508 SRR1039509 ... SRR1039520 SRR1039521
colData names(9): SampleName cell ... Sample BioSampleTo retrieve the experiment data from a SummarizedExperiment object one can use the assays() accessor. An object can have multiple assay datasets each of which can be accessed using the $ operator. The airway dataset contains only one assay (counts). Here each row represents a gene transcript and each column one of the samples.
assays(se)$counts| SRR1039508 | SRR1039509 | SRR1039512 | SRR1039513 | SRR1039516 | SRR1039517 | SRR1039520 | SRR1039521 | |
|---|---|---|---|---|---|---|---|---|
| ENSG00000000003 | 679 | 448 | 873 | 408 | 1138 | 1047 | 770 | 572 |
| ENSG00000000005 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| ENSG00000000419 | 467 | 515 | 621 | 365 | 587 | 799 | 417 | 508 |
| ENSG00000000457 | 260 | 211 | 263 | 164 | 245 | 331 | 233 | 229 |
| ENSG00000000460 | 60 | 55 | 40 | 35 | 78 | 63 | 76 | 60 |
| ENSG00000000938 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 |
| ENSG00000000971 | 3251 | 3679 | 6177 | 4252 | 6721 | 11027 | 5176 | 7995 |
| ENSG00000001036 | 1433 | 1062 | 1733 | 881 | 1424 | 1439 | 1359 | 1109 |
| ENSG00000001084 | 519 | 380 | 595 | 493 | 820 | 714 | 696 | 704 |
| ENSG00000001167 | 394 | 236 | 464 | 175 | 658 | 584 | 360 | 269 |
25.1.2 ‘Row’ (regions-of-interest) data
The rowRanges() accessor is used to view the range information for a RangedSummarizedExperiment. (Note if this were the parent SummarizedExperiment class we’d use rowData()). The data are stored in a GRangesList object, where each list element corresponds to one gene transcript and the ranges in each GRanges correspond to the exons in the transcript.
rowRanges(se)GRangesList object of length 63677:
$ENSG00000000003
GRanges object with 17 ranges and 2 metadata columns:
seqnames ranges strand | exon_id exon_name
<Rle> <IRanges> <Rle> | <integer> <character>
[1] X 99883667-99884983 - | 667145 ENSE00001459322
[2] X 99885756-99885863 - | 667146 ENSE00000868868
[3] X 99887482-99887565 - | 667147 ENSE00000401072
[4] X 99887538-99887565 - | 667148 ENSE00001849132
[5] X 99888402-99888536 - | 667149 ENSE00003554016
... ... ... ... . ... ...
[13] X 99890555-99890743 - | 667156 ENSE00003512331
[14] X 99891188-99891686 - | 667158 ENSE00001886883
[15] X 99891605-99891803 - | 667159 ENSE00001855382
[16] X 99891790-99892101 - | 667160 ENSE00001863395
[17] X 99894942-99894988 - | 667161 ENSE00001828996
-------
seqinfo: 722 sequences (1 circular) from an unspecified genome
...
<63676 more elements>25.1.3 ‘Column’ (sample) data
Sample meta-data describing the samples can be accessed using colData(), and is a DataFrame that can store any number of descriptive columns for each sample row.
colData(se)DataFrame with 8 rows and 9 columns
SampleName cell dex albut Run avgLength
<factor> <factor> <factor> <factor> <factor> <integer>
SRR1039508 GSM1275862 N61311 untrt untrt SRR1039508 126
SRR1039509 GSM1275863 N61311 trt untrt SRR1039509 126
SRR1039512 GSM1275866 N052611 untrt untrt SRR1039512 126
SRR1039513 GSM1275867 N052611 trt untrt SRR1039513 87
SRR1039516 GSM1275870 N080611 untrt untrt SRR1039516 120
SRR1039517 GSM1275871 N080611 trt untrt SRR1039517 126
SRR1039520 GSM1275874 N061011 untrt untrt SRR1039520 101
SRR1039521 GSM1275875 N061011 trt untrt SRR1039521 98
Experiment Sample BioSample
<factor> <factor> <factor>
SRR1039508 SRX384345 SRS508568 SAMN02422669
SRR1039509 SRX384346 SRS508567 SAMN02422675
SRR1039512 SRX384349 SRS508571 SAMN02422678
SRR1039513 SRX384350 SRS508572 SAMN02422670
SRR1039516 SRX384353 SRS508575 SAMN02422682
SRR1039517 SRX384354 SRS508576 SAMN02422673
SRR1039520 SRX384357 SRS508579 SAMN02422683
SRR1039521 SRX384358 SRS508580 SAMN02422677This sample metadata can be accessed using the $ accessor which makes it easy to subset the entire object by a given phenotype.
# subset for only those samples treated with dexamethasone
se[, se$dex == "trt"]class: RangedSummarizedExperiment
dim: 63677 4
metadata(1): ''
assays(1): counts
rownames(63677): ENSG00000000003 ENSG00000000005 ... ENSG00000273492
ENSG00000273493
rowData names(10): gene_id gene_name ... seq_coord_system symbol
colnames(4): SRR1039509 SRR1039513 SRR1039517 SRR1039521
colData names(9): SampleName cell ... Sample BioSample25.1.4 Experiment-wide metadata
Meta-data describing the experimental methods and publication references can be accessed using metadata().
metadata(se)[[1]]
Experiment data
Experimenter name: Himes BE
Laboratory: NA
Contact information:
Title: RNA-Seq transcriptome profiling identifies CRISPLD2 as a glucocorticoid responsive gene that modulates cytokine function in airway smooth muscle cells.
URL: http://www.ncbi.nlm.nih.gov/pubmed/24926665
PMIDs: 24926665
Abstract: A 226 word abstract is available. Use 'abstract' method.Note that metadata() is just a simple list, so it is appropriate for any experiment wide metadata the user wishes to save, such as storing model formulas.
metadata(se)$formula <- counts ~ dex + albut
metadata(se)[[1]]
Experiment data
Experimenter name: Himes BE
Laboratory: NA
Contact information:
Title: RNA-Seq transcriptome profiling identifies CRISPLD2 as a glucocorticoid responsive gene that modulates cytokine function in airway smooth muscle cells.
URL: http://www.ncbi.nlm.nih.gov/pubmed/24926665
PMIDs: 24926665
Abstract: A 226 word abstract is available. Use 'abstract' method.
$formula
counts ~ dex + albut
25.2 Common operations on SummarizedExperiment
25.2.1 Subsetting
-
[Performs two dimensional subsetting, just like subsetting a matrix or data frame.
# subset the first five transcripts and first three samples
se[1:5, 1:3]class: RangedSummarizedExperiment
dim: 5 3
metadata(2): '' formula
assays(1): counts
rownames(5): ENSG00000000003 ENSG00000000005 ENSG00000000419
ENSG00000000457 ENSG00000000460
rowData names(10): gene_id gene_name ... seq_coord_system symbol
colnames(3): SRR1039508 SRR1039509 SRR1039512
colData names(9): SampleName cell ... Sample BioSample-
$operates oncolData()columns, for easy sample extraction.
se[, se$cell == "N61311"]class: RangedSummarizedExperiment
dim: 63677 2
metadata(2): '' formula
assays(1): counts
rownames(63677): ENSG00000000003 ENSG00000000005 ... ENSG00000273492
ENSG00000273493
rowData names(10): gene_id gene_name ... seq_coord_system symbol
colnames(2): SRR1039508 SRR1039509
colData names(9): SampleName cell ... Sample BioSample25.2.2 Getters and setters
counts <- matrix(1:15, 5, 3, dimnames=list(LETTERS[1:5], LETTERS[1:3]))
dates <- SummarizedExperiment(assays=list(counts=counts),
rowData=DataFrame(month=month.name[1:5], day=1:5))
# Subset all January assays
dates[rowData(dates)$month == "January", ]class: SummarizedExperiment
dim: 1 3
metadata(0):
assays(1): counts
rownames(1): A
rowData names(2): month day
colnames(3): A B C
colData names(0):-
assay()versusassays()There are two accessor functions for extracting the assay data from aSummarizedExperimentobject.assays()operates on the entire list of assay data as a whole, whileassay()operates on only one assay at a time.assay(x, i)is simply a convenience function which is equivalent toassays(x)[[i]].
assays(se)List of length 1
names(1): countsassays(se)[[1]][1:5, 1:5] SRR1039508 SRR1039509 SRR1039512 SRR1039513 SRR1039516
ENSG00000000003 679 448 873 408 1138
ENSG00000000005 0 0 0 0 0
ENSG00000000419 467 515 621 365 587
ENSG00000000457 260 211 263 164 245
ENSG00000000460 60 55 40 35 78# assay defaults to the first assay if no i is given
assay(se)[1:5, 1:5] SRR1039508 SRR1039509 SRR1039512 SRR1039513 SRR1039516
ENSG00000000003 679 448 873 408 1138
ENSG00000000005 0 0 0 0 0
ENSG00000000419 467 515 621 365 587
ENSG00000000457 260 211 263 164 245
ENSG00000000460 60 55 40 35 78assay(se, 1)[1:5, 1:5] SRR1039508 SRR1039509 SRR1039512 SRR1039513 SRR1039516
ENSG00000000003 679 448 873 408 1138
ENSG00000000005 0 0 0 0 0
ENSG00000000419 467 515 621 365 587
ENSG00000000457 260 211 263 164 245
ENSG00000000460 60 55 40 35 7825.2.3 Range-based operations
-
subsetByOverlaps()SummarizedExperimentobjects support all of thefindOverlaps()methods and associated functions. This includessubsetByOverlaps(), which makes it easy to subset aSummarizedExperimentobject by an interval.
In tne next code block, we define a region of interest (or many regions of interest) and then subset our SummarizedExperiment by overlaps with this region.
# Subset for only rows which are in the interval 100,000 to 110,000 of
# chromosome 1
roi <- GRanges(seqnames="1", ranges=100000:1100000)
sub_se = subsetByOverlaps(se, roi)
sub_seclass: RangedSummarizedExperiment
dim: 74 8
metadata(2): '' formula
assays(1): counts
rownames(74): ENSG00000131591 ENSG00000177757 ... ENSG00000272512
ENSG00000273443
rowData names(10): gene_id gene_name ... seq_coord_system symbol
colnames(8): SRR1039508 SRR1039509 ... SRR1039520 SRR1039521
colData names(9): SampleName cell ... Sample BioSampledim(sub_se)[1] 74 8
25.3 Constructing a SummarizedExperiment
To construct a SummarizedExperiment object, you need to provide the following components: - assays: A list of matrices or matrix-like objects containing the data. - rowData: A DataFrame containing information about the features (rows). - colData: A DataFrame containing information about the samples (columns). - +/- metadata: A list containing additional metadata about the experiment.
For a nearly real example, we will use the DeRisi dataset. We’ll start with the original data, which is a data.frame with the first couple of columns containing the gene information and the rest of the columns containing the data.
# Load the DeRisi dataset
deRisi <- read.csv("https://raw.githubusercontent.com/seandavi/RBiocBook/refs/heads/main/data/derisi.csv")
head(deRisi) ORF Name R1 R2 R3 R4 R5 R6 R7 R1.Bkg R2.Bkg R3.Bkg
1 YHR007C ERG11 7896 7484 14679 14617 9853 7599 6490 1155 1984 1323
2 YBR218C PYC2 12144 11177 10241 4820 4950 7047 17035 1074 1694 1243
3 YAL051W FUN43 4478 6435 6230 6848 5111 7180 4497 1140 1950 1649
4 YAL053W 6343 8243 6743 3304 3556 4694 3849 1020 1897 1196
5 YAL054C ACS1 1542 3044 2076 1695 1753 4806 10802 1082 1940 1504
6 YAL055W 1769 3243 2094 1367 1853 3580 1956 975 1821 1185
R4.Bkg R5.Bkg R6.Bkg R7.Bkg G1 G2 G3 G4 G5 G6 G7 G1.Bkg
1 1171 914 2445 981 8432 7173 11736 16798 12315 16111 13931 2404
2 876 1211 2444 742 11509 10226 13372 6500 6255 9024 6904 2148
3 1183 898 2637 927 5865 5895 5345 6302 5400 7933 5026 2422
4 881 1045 2518 697 6762 7454 6323 3595 4689 5660 4145 2107
5 1108 902 2610 980 3138 3785 2419 2114 2763 3561 1897 2405
6 851 1047 2536 698 2844 4069 2583 1651 2530 3484 1550 1674
G2.Bkg G3.Bkg G4.Bkg G5.Bkg G6.Bkg G7.Bkg
1 2561 1598 1506 1696 2667 1244
2 2527 1641 1196 1553 2569 848
3 2496 1902 1501 1644 2808 1154
4 2663 1607 1162 1577 2544 857
5 2528 1847 1445 1713 2767 1142
6 2648 1591 1114 1528 2668 870To convert this to a SummarizedExperiment, we need to extract the assay data, row data, and column data. The assay data will be the numeric values in the data frame, the row data will be the gene information, and the column data will be the sample information.
25.3.1 rowData, or feature information
Let’s start with the rowData, which will be a DataFrame containing the gene information. We can use the first two columns of the data frame for this purpose.
rdata <- deRisi[, 1:2]
head(rdata) ORF Name
1 YHR007C ERG11
2 YBR218C PYC2
3 YAL051W FUN43
4 YAL053W
5 YAL054C ACS1
6 YAL055W 25.3.2 colData, or sample information
Next, we will create the colData, which will be a DataFrame containing the sample information. Since the sample information really isn’t in the dataset, we will create a simple DataFrame with sample names.
cdata <- DataFrame(sample=paste("Sample", 0:6), timepoint = 0:6,
hours = c(0, 9.5,11.5,13.5,15.5,18.5,20.5))
head(cdata)DataFrame with 6 rows and 3 columns
sample timepoint hours
<character> <integer> <numeric>
1 Sample 0 0 0.0
2 Sample 1 1 9.5
3 Sample 2 2 11.5
4 Sample 3 3 13.5
5 Sample 4 4 15.5
6 Sample 5 5 18.525.3.3 assays, or the data
Remember that the DeRisi dataset has four different assays,
| assay | description |
|---|---|
| R | Red fluorescence |
| G | Green luorescence |
| Rb | Red background fluorescence |
| Gb | Green background fluorescence |
We will create a list of matrices, one for each assay. The matrices will be the numeric values in the data frame, excluding the first two columns.
When we create a SummarizedExperiment object, the “constructor” will check to see that the colnames of the matrices in the list are the same as the rownames of the colData DataFrame, and that the rownames of the matrices in the list are the same as the rownames of the rowData DataFrame.
So, we need to fix that all up. Let’s start wit the rownames of the rowData DataFrame:
rownames(rdata) <- rdata$ORFNow, let’s set the rownames of the coldata DataFrame to the sample names:
rownames(cdata) <- cdata$sampleNow, we can fix the rownames and colnames of the matrices for our R, G, Rb, and Gb assays:
Take a look at the matrices to make sure they look right.
25.3.4 Putting it all together
se <- SummarizedExperiment(assays=list(R=R, G=G, Rb=Rb, Gb=Gb), rowData=rdata, colData=cdata)25.3.5 Getting logRatios
Now that we have a SummarizedExperiment object, we can easily compute the log ratios of the Red and Green foreground fluorescence. This is a common operation in microarray data analysis.
Warning: NaNs producedassays(se)$logRatios <- logRatiosNow, we’ve added a new assay to the SummarizedExperiment object called logRatios. This assay contains the log ratios of the Red and Green foreground fluorescence.
assays(se)List of length 5
names(5): R G Rb Gb logRatiosAnd if we want to access the log ratios, we can do so using the assay() method:
Sample 0 Sample 1 Sample 2 Sample 3 Sample 4 Sample 5
YHR007C -1.2204686 NaN NaN 2.70275677 1.6545902 5.260867
YBR218C NaN NaN 2.9757832 2.39231742 1.9319816 3.983313
YAL051W 0.1135715 NaN NaN NaN -1.3681061 2.138654
YAL053W -1.3753298 NaN NaN 0.05044902 1.0906497 5.215440
YAL054C 0.2706476 0.333657387 0.0000000 0.31420165 0.3165815 NaN
YAL055W 0.6209723 -0.001745548 0.2683547 0.11082813 0.4931189 NaN
Sample 6
YHR007C 4.8223618
YBR218C NaN
YAL051W 1.2205754
YAL053W 0.8875253
YAL054C NaN
YAL055W NaN Sample 0 Sample 1 Sample 2 Sample 3 Sample 4 Sample 5
YHR007C -1.2204686 NaN NaN 2.70275677 1.6545902 5.260867
YBR218C NaN NaN 2.9757832 2.39231742 1.9319816 3.983313
YAL051W 0.1135715 NaN NaN NaN -1.3681061 2.138654
YAL053W -1.3753298 NaN NaN 0.05044902 1.0906497 5.215440
YAL054C 0.2706476 0.333657387 0.0000000 0.31420165 0.3165815 NaN
YAL055W 0.6209723 -0.001745548 0.2683547 0.11082813 0.4931189 NaN
Sample 6
YHR007C 4.8223618
YBR218C NaN
YAL051W 1.2205754
YAL053W 0.8875253
YAL054C NaN
YAL055W NaN