Chapter 9 Phylogenetic data
(Rambaut et al. 2020)
(Singer et al. 2020)
## Warning in with_tz(Sys.time(), tzone): Unrecognized time zone ''see See Figure ??
groups = str_match(ntr$tip.label,'.*_([A-Z][\\.]?[0-9]?[0-9]?).*')
ntr = groupOTU(ntr, split(ntr$tip.label,groups[,2]))
ggtree(ntr, aes(color=group), layout="circular", branch.length="none") +
geom_tiplab(size=3, aes(angle=angle)) +
ggtitle('COVID Glue Phylogenetic Clades')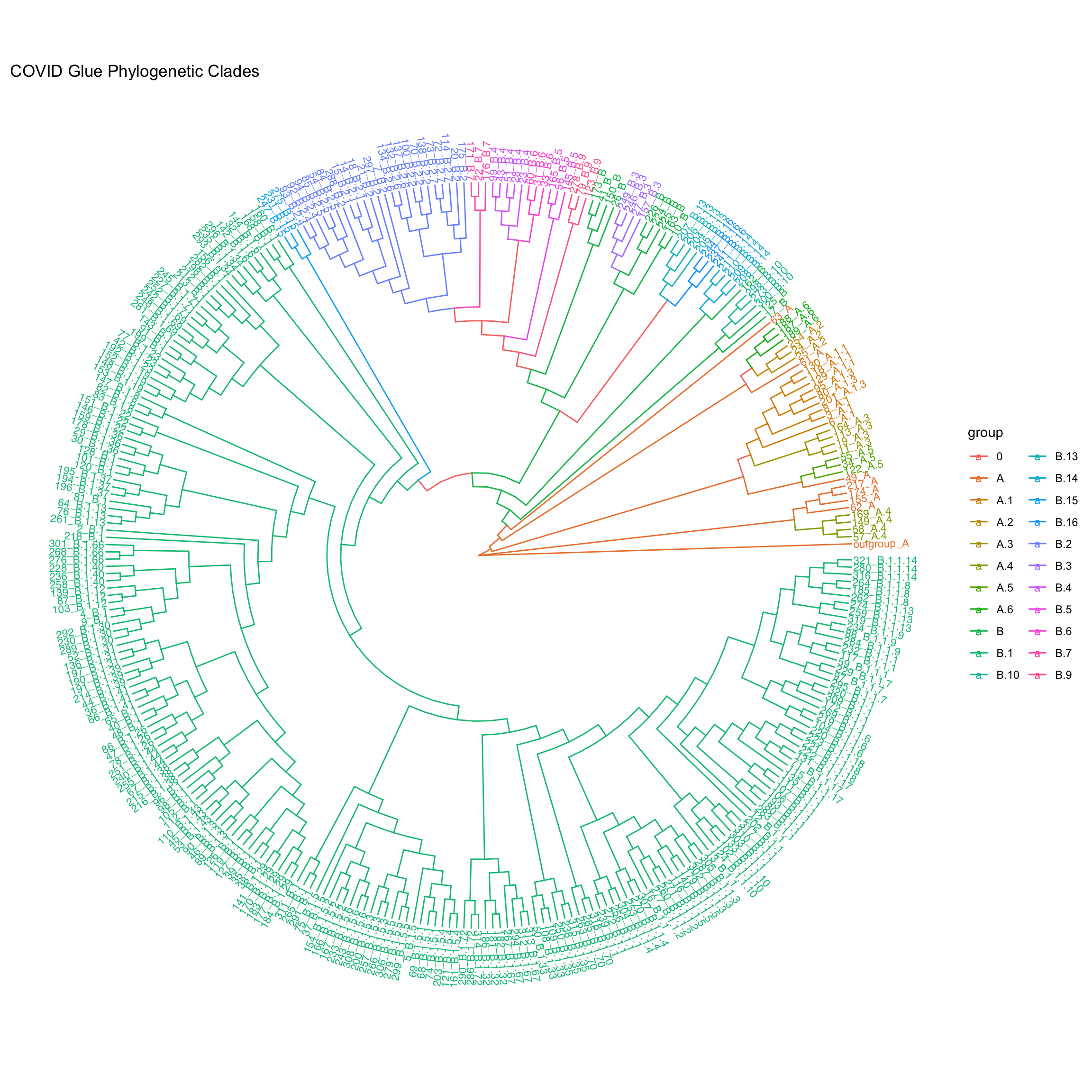
abc
dat = cov_glue_lineage_data()
dat = dat %>%
dplyr::mutate(lineage = sub('^([^.][.]?[^.]+).*', '\\1', lineage)) %>%
dplyr::mutate(region = countrycode::countrycode(country,origin='country.name',
destination='region')) %>%
dplyr::group_by(epiweek,lineage,region) %>%
dplyr::summarize(cases=n()) %>%
dplyr::filter(!is.na(region)) %>%
dplyr::ungroup()## `summarise()` regrouping output by 'epiweek', 'lineage' (override with `.groups` argument)## # A tibble: 6 x 4
## epiweek lineage region cases
## <dbl> <chr> <chr> <int>
## 1 0 B East Asia & Pacific 2
## 2 1 B East Asia & Pacific 20
## 3 2 A East Asia & Pacific 3
## 4 2 B East Asia & Pacific 7
## 5 3 A East Asia & Pacific 9
## 6 3 B East Asia & Pacific 10p = dat %>%
ggplot(aes(x=epiweek,y=cases,fill=lineage)) +
geom_bar(stat='identity', position='fill') +
facet_wrap("region",ncol=2) +
theme(legend.position='bottom')
p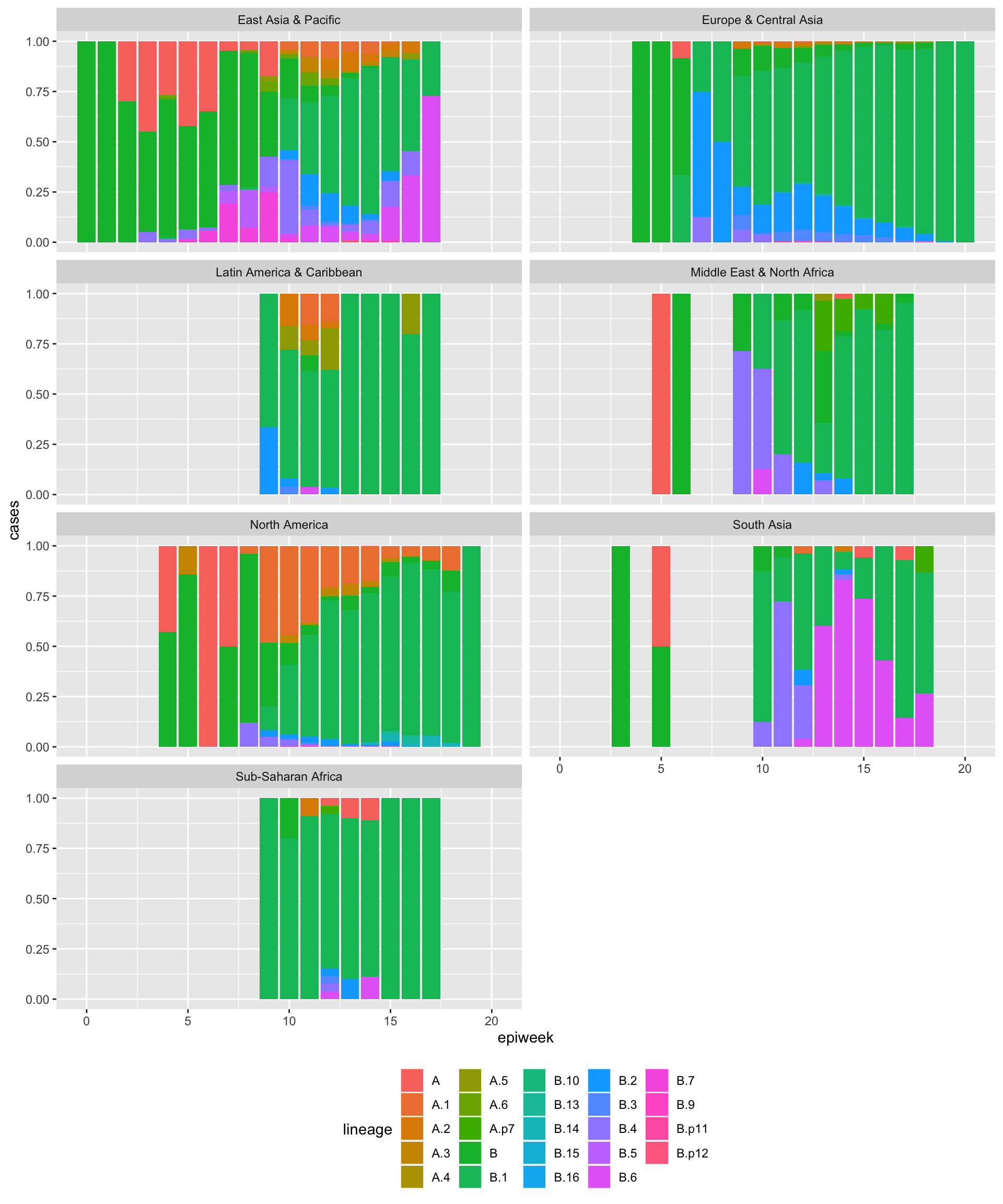
9.1 Notes
Rambaut, Andrew, Edward C. Holmes, Áine O’Toole, Verity Hill, John T. McCrone, Christopher Ruis, Louis du Plessis, and Oliver G. Pybus. 2020. “A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology.” Nat. Microbiol. 5 (November): 1403–7. https://doi.org/10.1038/s41564-020-0770-5.
Singer, Joshua, Robert Gifford, Matthew Cotten, and David Robertson. 2020. “CoV-GLUE: A Web Application for Tracking SARS-CoV-2 Genomic Variation.” Preprints, June. https://doi.org/10.20944/preprints202006.0225.v1.
References
Rambaut, Andrew, Edward C. Holmes, Áine O’Toole, Verity Hill, John T. McCrone, Christopher Ruis, Louis du Plessis, and Oliver G. Pybus. 2020. “A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology.” Nat. Microbiol. 5 (November): 1403–7. https://doi.org/10.1038/s41564-020-0770-5.
Singer, Joshua, Robert Gifford, Matthew Cotten, and David Robertson. 2020. “CoV-GLUE: A Web Application for Tracking SARS-CoV-2 Genomic Variation.” Preprints, June. https://doi.org/10.20944/preprints202006.0225.v1.