While the inducedSubgraphByPkgs()
returns the subgraph with the minimal connections
between named packages, this function takes a vector of
package names, a degree (1 or more) and returns the
subgraph(s) that are within degree of the
package named.
subgraphByDegree(g, pkg, degree = 1, ...)Arguments
- g
an igraph graph, typically created by
buildPkgDependencyIgraph()- pkg
character(1)package name from which to measure degree.- degree
integer(1) degree, limit search for adjacent vertices to this degree.
- ...
passed on to
igraph::distances()
Value
An igraph graph, with only nodes and their edges within degree of the named package
Examples
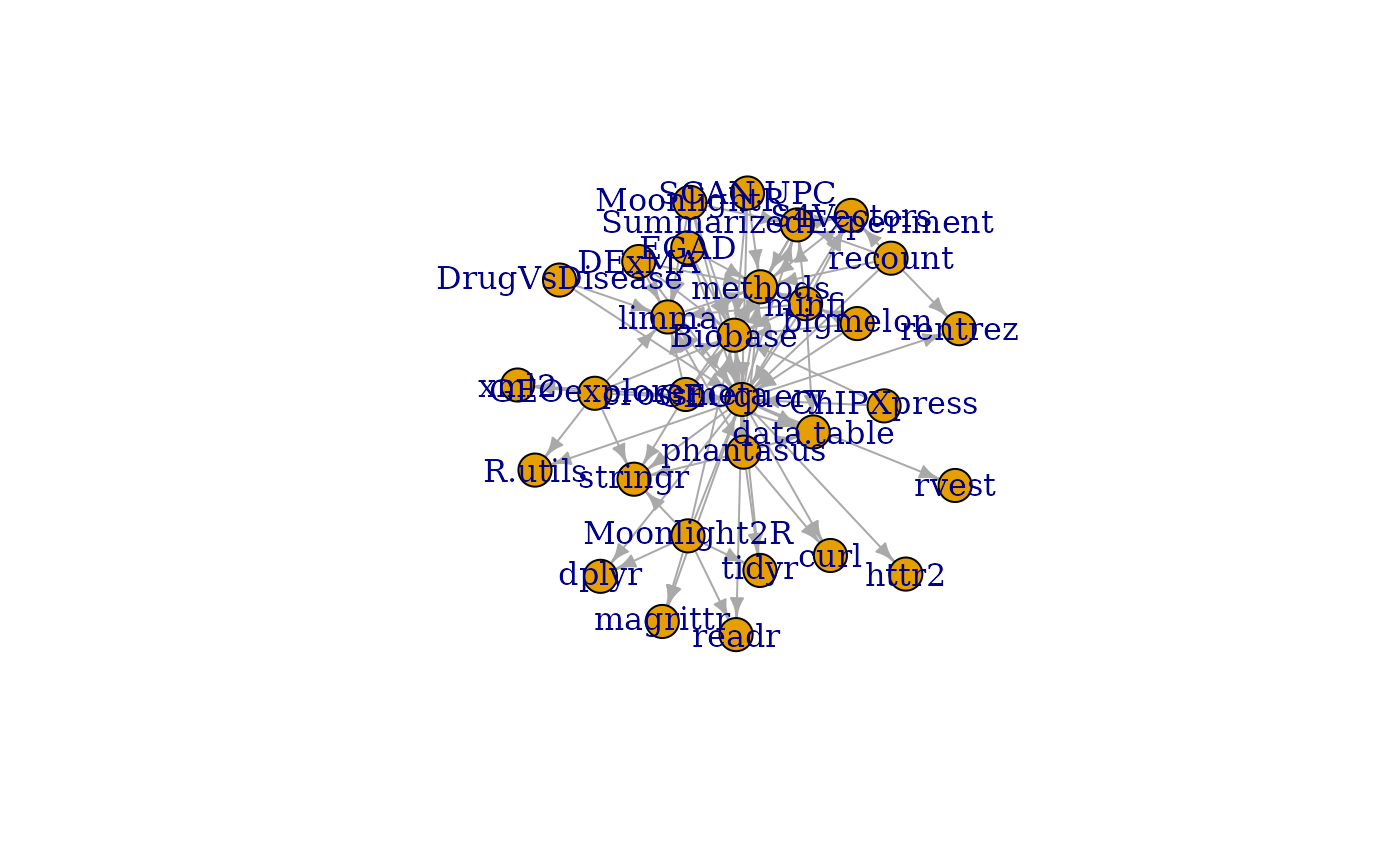
g <- buildPkgDependencyIgraph(buildPkgDependencyDataFrame())
#> 'getOption("repos")' replaces Bioconductor standard repositories, see
#> 'help("repositories", package = "BiocManager")' for details.
#> Replacement repositories:
#> CRAN: https://cran.rstudio.com
g2 <- subgraphByDegree(g, 'GEOquery')
plot(g2)