Using the SRAdbV2 Package
Sean Davis
Center for Cancer Research, National Cancer Institute, NIH, Bethesda, MD USAseandavi@gmail.com
SRAdbv2.RmdInstallation
The SRAdbV2 package is currently available from GitHub and is under active development. Either the devtools package or the BiocManager package can be used for easy installation.
Usage
The Omicidx
The entrypoint for using the SRAdbV2 system is the Omicidx, an R6 class. To start, create a new instance of the class.
Typing oidx$ and then TAB will give possible completions. Note the “search” completion.
Queries
Once an instance of Omicidx is created (here we will call the instance oidx), search capabilities are available via oidx$search(). The one interesting parameter is the q parameter. This parameter takes a string formatted as a Lucene query string. See below for Query syntax.
query=paste(
paste0('sample.taxon_id:', 10116),
'AND experiment.library_strategy:"rna seq"',
'AND experiment.library_source:transcriptomic',
'AND experiment.platform:illumina')
z = oidx$search(q=query,entity='full',size=100L)The entity parameter is one of the SRA entity types available via the API. The size parameter is the number of records that will be returned in each “chunk”.
Fetching results
Because result sets can be large, we have a special method that allows us to “scroll” through the results or to simply get them en bloc. The first step for result retrieval, then, is to get a Scroller.
## <Scroller>
## Public:
## clone: function (deep = FALSE)
## collate: function (limit = Inf)
## count: active binding
## fetched: active binding
## has_next: function ()
## initialize: function (search, progress = interactive())
## reset: function ()
## yield: function ()
## Private:
## .count: NULL
## .fetched: 0
## .last: FALSE
## progress: FALSE
## scroll: 1m
## scroll_id: NULL
## search: Searcher, R6Methods such as s$count allow introspection into the available number of results, in this case, 10221 records.
The Scroller provides two different approaches to accessing the resulting data.
Collating entire result sets
The first approach to getting results of a query back into R is the most convenient, but for large result sets, the entire dataset is loaded into memory and may take significant time if network connections are slow.
# for VERY large result sets, this may take
# quite a bit of time and/or memory. An
# alternative is to use s$chunk() to retrieve
# one batch of records at a time and process
# incrementally.
res = s$collate(limit = 1000)
head(res)## # A tibble: 6 x 83
## experiment.Insdc experiment.LastMet… experiment.LastUpd…
## <lgl> <dttm> <dttm>
## 1 TRUE 2013-09-23 16:45:14 2013-09-23 16:45:14
## 2 TRUE 2013-09-23 16:45:14 2013-09-23 16:45:14
## 3 TRUE 2017-09-24 06:21:35 2017-09-28 08:48:29
## 4 TRUE 2017-09-24 06:21:34 2017-09-28 08:43:12
## 5 TRUE 2017-09-24 06:21:37 2017-09-28 08:55:15
## 6 TRUE 2017-09-24 06:21:36 2017-09-28 08:50:14
## # ... with 80 more variables: experiment.Published <dttm>,
## # experiment.Received <dttm>, experiment.Status <chr>,
## # experiment.accession <chr>, experiment.alias <chr>,
## # experiment.attributes <list>, experiment.center_name <chr>,
## # experiment.experiment_accession <chr>, experiment.identifiers <list>,
## # experiment.instrument_model <chr>, experiment.library_layout <chr>,
## # experiment.library_name <chr>, experiment.library_selection <chr>,
## # experiment.library_source <chr>, experiment.library_strategy <chr>,
## # experiment.platform <chr>, experiment.sample_accession <chr>,
## # experiment.study_accession <chr>, experiment.title <chr>,
## # experiment.xrefs <list>, experiment.broker_name <chr>,
## # experiment.design <chr>,
## # experiment.library_construction_protocol <chr>,
## # experiment.library_layout_length <dbl>,
## # experiment.library_layout_sdev <chr>, sample.BioSample <chr>,
## # sample.GEO <chr>, sample.Insdc <lgl>, sample.LastMetaUpdate <dttm>,
## # sample.LastUpdate <dttm>, sample.Published <dttm>,
## # sample.Received <dttm>, sample.Status <chr>, sample.accession <chr>,
## # sample.alias <chr>, sample.attributes <list>,
## # sample.center_name <chr>, sample.identifiers <list>,
## # sample.organism <chr>, sample.taxon_id <int>, sample.title <chr>,
## # sample.xrefs <list>, sample.description <chr>, study.BioProject <chr>,
## # study.GEO <chr>, study.Insdc <lgl>, study.LastMetaUpdate <dttm>,
## # study.LastUpdate <dttm>, study.Published <dttm>,
## # study.Received <dttm>, study.Status <chr>, study.abstract <chr>,
## # study.accession <chr>, study.alias <chr>, study.center_name <chr>,
## # study.identifiers <list>, study.study_accession <chr>,
## # study.study_type <chr>, study.title <chr>, study.attributes <list>,
## # study.broker_name <chr>, study.description <chr>, study.xrefs <list>,
## # Insdc <lgl>, LastMetaUpdate <dttm>, LastUpdate <dttm>,
## # Published <dttm>, Received <dttm>, Status <chr>, accession <chr>,
## # alias <chr>, attributes <list>, center_name <chr>, identifiers <list>,
## # reads <list>, spot_length <int>, broker_name <chr>, run_center <chr>,
## # nreads <int>, sample.broker_name <chr>Note that the scroller now reports that it has fetched (s$fetched) 1000 records.
To reuse a Scroller, we must reset it first.
## <Scroller>
## Public:
## clone: function (deep = FALSE)
## collate: function (limit = Inf)
## count: active binding
## fetched: active binding
## has_next: function ()
## initialize: function (search, progress = interactive())
## reset: function ()
## yield: function ()
## Private:
## .count: 10221
## .fetched: 0
## .last: FALSE
## progress: FALSE
## scroll: 1m
## scroll_id: NULL
## search: Searcher, R6Yielding chunks
The second approach is to iterate through results using the yield method. This approach allows the user to perform processing on chunks of data as they arrive in R.
j = 0
## fetch only 500 records, but
## `yield` will return NULL
## after ALL records have been fetched
while(s$fetched < 500) {
res = s$yield()
# do something interesting with `res` here if you like
j = j + 1
message(sprintf('total of %d fetched records, loop iteration # %d', s$fetched, j))
}## total of 100 fetched records, loop iteration # 1## total of 200 fetched records, loop iteration # 2## total of 300 fetched records, loop iteration # 3## total of 400 fetched records, loop iteration # 4## total of 500 fetched records, loop iteration # 5The Scroller also has a has_next() method that will report TRUE if the result set has not been fully fetched. Using the reset() method will move the cursor back to the beginning of the result set.
Query syntax
Terms
A query is broken up into terms and operators. There are two types of terms: Single Terms and Phrases. A Single Term is a single word such as “test” or “hello”. A Phrase is a group of words surrounded by double quotes such as “hello dolly”. Multiple terms can be combined together with Boolean operators to form a more complex query (see below).
Fields
Queries support fielded data. When performing a search you can either specify a field, or use the default field. The field names and default field is implementation specific. You can search any field by typing the field name followed by a colon “:” and then the term you are looking for. As an example, let’s assume a Lucene index contains two fields, title and abstract. If you want to find the document entitled “The Right Way” which contains the text “don’t go this way” in the abstract, you can enter:
title:"The Right Way" AND abstract:goor
Note: The field is only valid for the term that it directly precedes, so the query
title:Do it rightwill only find “Do” in the title field. It will find “it” and “right” in any other fields.
Wildcard Searches
Lucene supports single and multiple character wildcard searches within single terms (not within phrase queries). To perform a single character wildcard search use the “?” symbol. To perform a multiple character wildcard search use the “*" symbol. The single character wildcard search looks for terms that match that with the single character replaced. For example, to search for “text” or “test” you can use the search:
te?tMultiple character wildcard searches looks for 0 or more characters. For example, to search for test, tests or tester, you can use the search:
test*You can also use the wildcard searches in the middle of a term.
te*tNote: You cannot use a * or ? symbol as the first character of a search.
Fuzzy Searches
Lucene supports fuzzy searches based on the Levenshtein Distance, or Edit Distance algorithm. To do a fuzzy search use the tilde, “~”, symbol at the end of a Single word Term. For example to search for a term similar in spelling to “roam” use the fuzzy search:
roam~This search will find terms like foam and roams.
Starting with Lucene 1.9 an additional (optional) parameter can specify the required similarity. The value is between 0 and 1, with a value closer to 1 only terms with a higher similarity will be matched. For example:
roam~0.8The default that is used if the parameter is not given is 0.5.
Proximity Searches
Lucene supports finding words are a within a specific distance away. To do a proximity search use the tilde, “~”, symbol at the end of a Phrase. For example to search for a “apache” and “jakarta” within 10 words of each other in a document use the search:
"jakarta apache"~10Range Searches
Range Queries allow one to match documents whose field(s) values are between the lower and upper bound specified by the Range Query. Range Queries can be inclusive or exclusive of the upper and lower bounds. Sorting is done lexicographically.
mod_date:[20020101 TO 20030101]This will find documents whose mod_date fields have values between 20020101 and 20030101, inclusive. Note that Range Queries are not reserved for date fields. You could also use range queries with non-date fields:
title:{Aida TO Carmen}This will find all documents whose titles are between Aida and Carmen, but not including Aida and Carmen. Inclusive range queries are denoted by square brackets. Exclusive range queries are denoted by curly brackets.
Boolean Operators
Boolean operators allow terms to be combined through logic operators. Lucene supports AND, “+”, OR, NOT and “-” as Boolean operators(Note: Boolean operators must be ALL CAPS).
OR
The OR operator is the default conjunction operator. This means that if there is no Boolean operator between two terms, the OR operator is used. The OR operator links two terms and finds a matching document if either of the terms exist in a document. This is equivalent to a union using sets. The symbol || can be used in place of the word OR.
To search for documents that contain either “jakarta apache” or just “jakarta” use the query:
"jakarta apache" jakartaor
"jakarta apache" OR jakartaThe AND operator matches documents where both terms exist anywhere in the text of a single document. This is equivalent to an intersection using sets. The symbol && can be used in place of the word AND. To search for documents that contain “jakarta apache” and “Apache Lucene” use the query:
“jakarta apache” AND “Apache Lucene”
+
The “+” or required operator requires that the term after the “+” symbol exist somewhere in a the field of a single document. To search for documents that must contain “jakarta” and may contain “lucene” use the query:
+jakarta luceneNOT
The NOT operator excludes documents that contain the term after NOT. This is equivalent to a difference using sets. The symbol ! can be used in place of the word NOT. To search for documents that contain “jakarta apache” but not “Apache Lucene” use the query:
"jakarta apache" NOT "Apache Lucene"Note: The NOT operator cannot be used with just one term. For example, the following search will return no results:
NOT "jakarta apache"Grouping
Lucene supports using parentheses to group clauses to form sub queries. This can be very useful if you want to control the boolean logic for a query. To search for either “jakarta” or “apache” and “website” use the query:
(jakarta OR apache) AND websiteThis eliminates any confusion and makes sure you that website must exist and either term jakarta or apache may exist.
Lucene supports using parentheses to group multiple clauses to a single field.
To search for a title that contains both the word “return” and the phrase “pink panther” use the query:
title:(+return +"pink panther")Escaping Special Characters
Lucene supports escaping special characters that are part of the query syntax. The current list special characters are
+ - && || ! ( ) { } [ ] ^ " ~ * ? : \To escape these character use the before the character. For example to search for (1+1):2 use the query:
\(1\+1\)\:2Using the raw API without R/Bioconductor
The SRAdbV2 is a client to a high-performance web-based API. As such, the web API is perfectly usable from either a simple test page, accessible here:
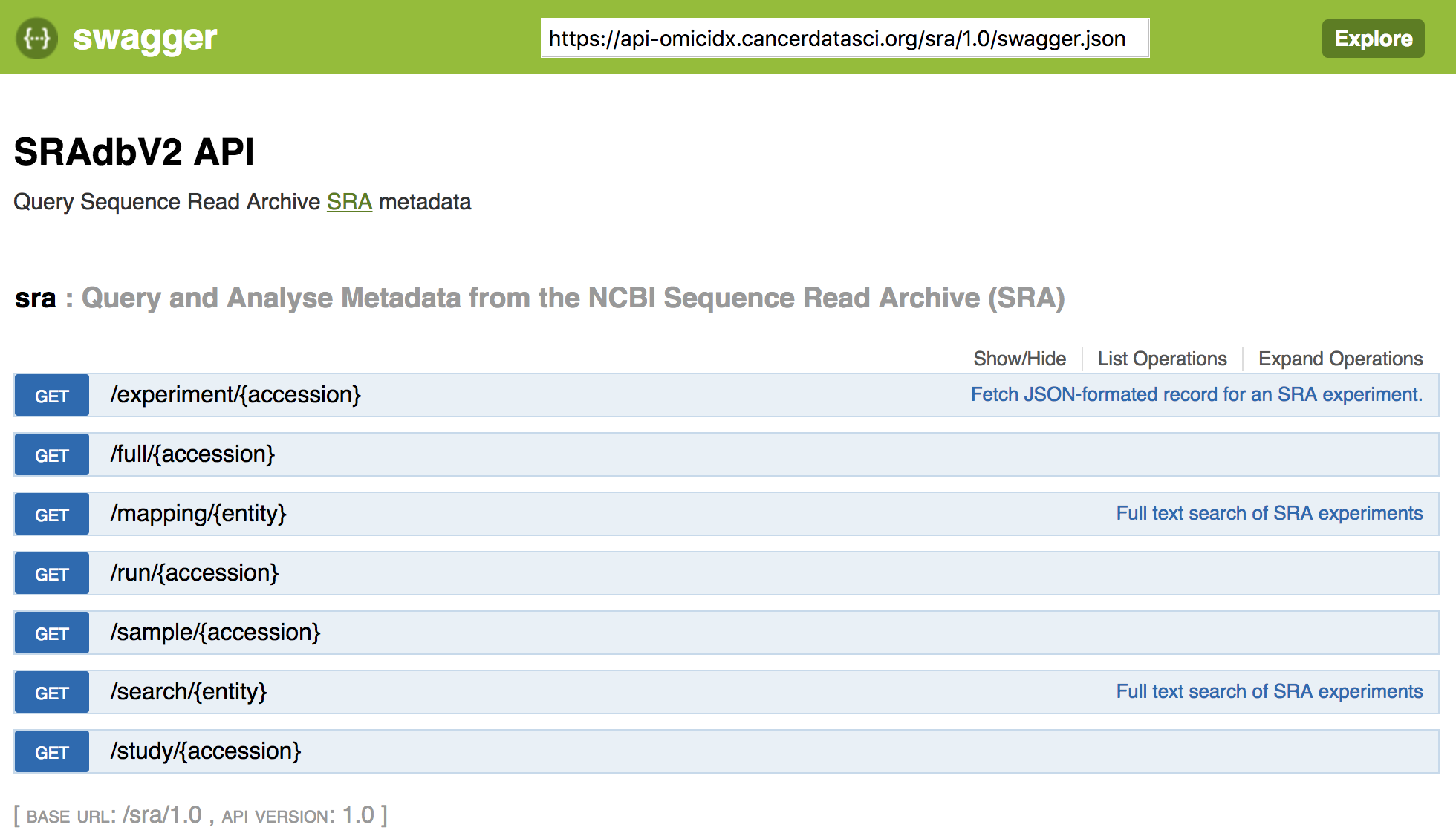
The web-based API provides a useful interface for experiment with queries. It also returns URLs associated with the example queries, facilitating querying with other tools like curl or wget.
The API is described using the OpenAPI standard, also known as Swagger. Tooling exists to quickly scaffold clients in any language (basically) based on the json available here:
## [1] "https://api-omicidx.cancerdatasci.org/sra/1.0/swagger.json"Provenance
## R version 3.5.1 (2018-07-02)
## Platform: x86_64-apple-darwin15.6.0 (64-bit)
## Running under: macOS Sierra 10.12.6
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] SRAdbV2_0.7.2 R6_2.3.0 BiocStyle_2.8.2
##
## loaded via a namespace (and not attached):
## [1] Rcpp_0.12.19 compiler_3.5.1 pillar_1.3.0
## [4] bindr_0.1.1 tools_3.5.1 digest_0.6.18
## [7] jsonlite_1.5 lubridate_1.7.4 evaluate_0.12
## [10] memoise_1.1.0 tibble_1.4.2 pkgconfig_2.0.2
## [13] rlang_0.3.0.1 cli_1.0.1 commonmark_1.5
## [16] curl_3.2 yaml_2.2.0 pkgdown_1.1.0.9000
## [19] xfun_0.3 bindrcpp_0.2.2 stringr_1.3.1
## [22] httr_1.3.1 dplyr_0.7.7 roxygen2_6.1.0
## [25] xml2_1.2.0 knitr_1.20 desc_1.2.0
## [28] fs_1.2.6 rprojroot_1.3-2 tidyselect_0.2.5
## [31] glue_1.3.0 fansi_0.4.0 rmarkdown_1.10
## [34] bookdown_0.7 purrr_0.2.5 magrittr_1.5
## [37] backports_1.1.2 htmltools_0.3.6 MASS_7.3-50
## [40] assertthat_0.2.0 utf8_1.1.4 stringi_1.2.4
## [43] crayon_1.3.4