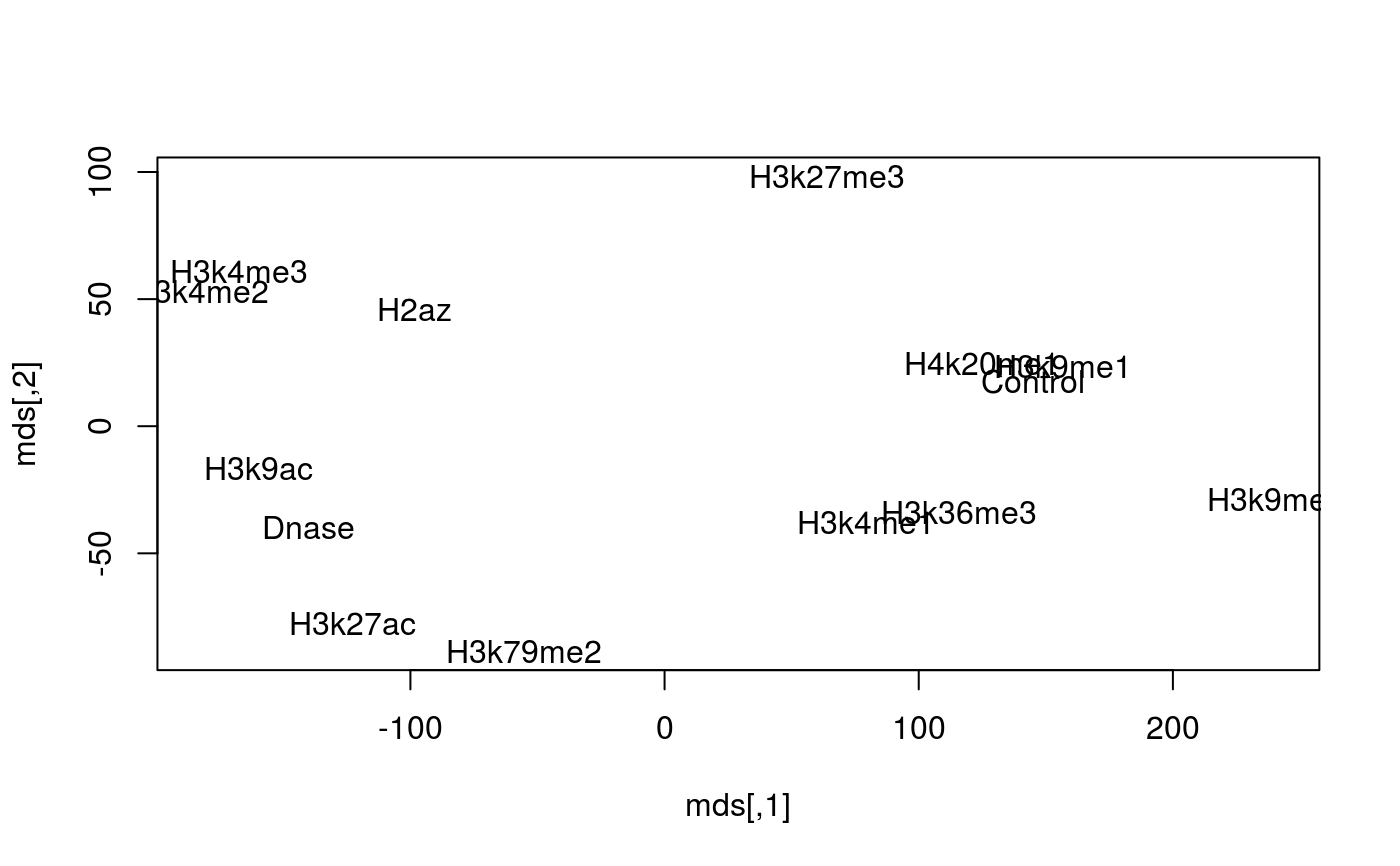
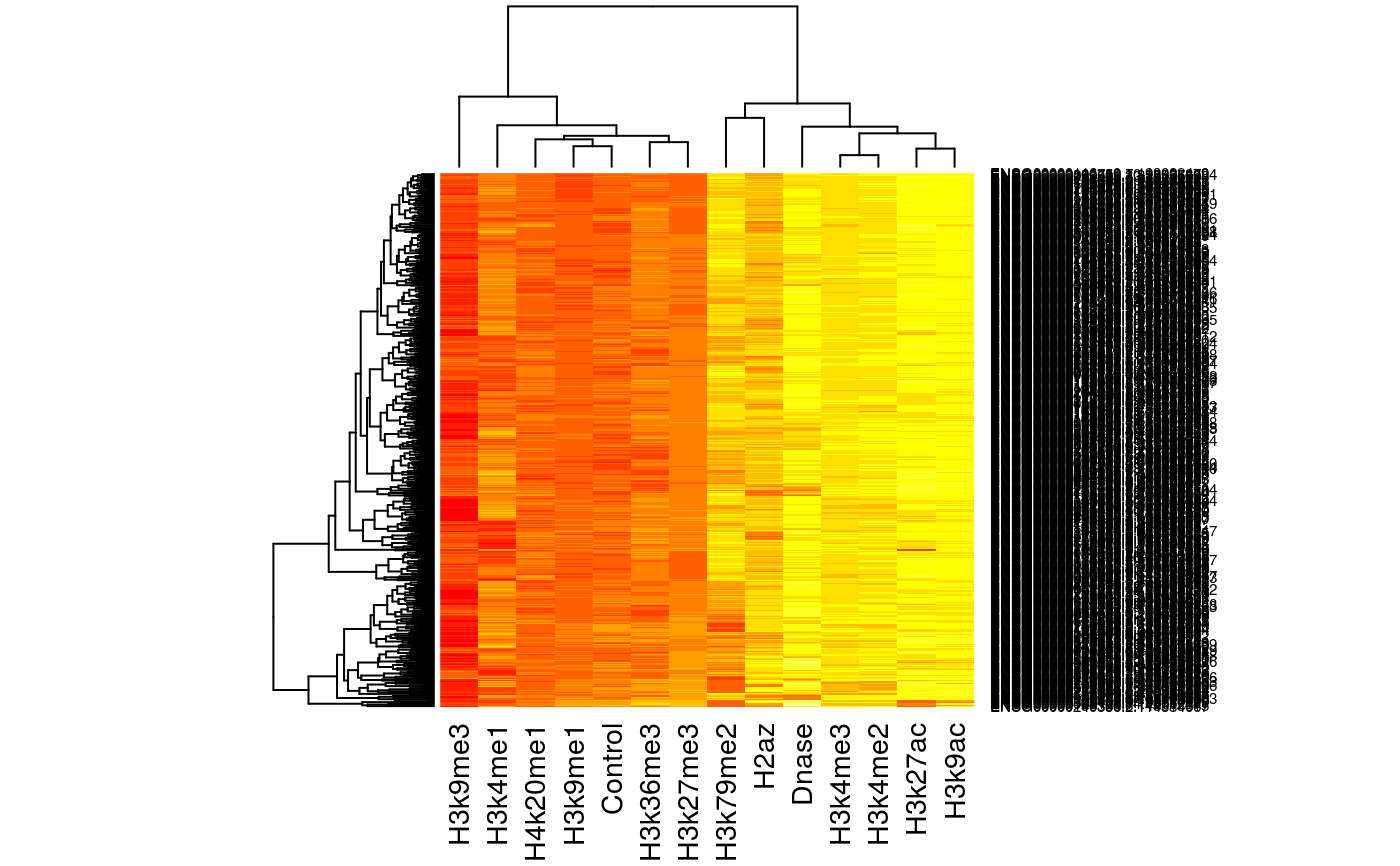
Load histone mark features, predictors
get_features.RdThis is a helper function that simply loads the the histone marks, one row per gene. Marks are given as column names. Row names are some strange mix of Ensembl IDs and junk.
get_features()
Value
a data.frame with histone marks in columns and one row per gene. Gene names are row names.
References
http://starbuck1.s3.amazonaws.com/expression-prediction/features.txt
Examples
feat = get_features() head(feat)#> Control Dnase H2az H3k27ac H3k27me3 #> ENSG00000000419.7.49575069 1.4889005 5.3014825 5.091915 5.9002487 1.926864 #> ENSG00000000457.8.169863093 2.4988150 5.9743785 3.938031 4.9268680 1.952509 #> ENSG00000000938.7.27961645 0.5709902 -0.3187665 1.623319 0.5011164 2.398604 #> ENSG00000001460.12.24740230 1.8854298 7.3455456 4.549188 5.2283188 2.345497 #> ENSG00000001461.11.24742304 1.3106251 7.3984600 5.530642 5.0248829 2.013595 #> ENSG00000001497.11.64754655 1.4852619 6.0811868 4.578087 5.1309106 1.827194 #> H3k36me3 H3k4me1 H3k4me2 H3k4me3 H3k79me2 #> ENSG00000000419.7.49575069 2.3510805 2.0440715 6.279558 5.770302 6.1590053 #> ENSG00000000457.8.169863093 2.0373866 3.3312018 5.320028 5.086961 4.4130180 #> ENSG00000000938.7.27961645 0.3580219 0.4790004 3.911917 3.184466 -0.8839223 #> ENSG00000001460.12.24740230 0.9295959 4.3682353 5.897041 5.730867 3.6809648 #> ENSG00000001461.11.24742304 1.9512642 4.4400132 6.577121 5.813670 4.6438269 #> ENSG00000001497.11.64754655 1.9878892 2.2879680 5.727750 5.253145 5.7361040 #> H3k9ac H3k9me1 H3k9me3 H4k20me1 #> ENSG00000000419.7.49575069 6.357470 0.8831567 1.2274619 2.9610552 #> ENSG00000000457.8.169863093 4.949883 1.0052250 1.0225077 0.8776918 #> ENSG00000000938.7.27961645 1.605415 1.1677089 -1.8420669 -0.6247215 #> ENSG00000001460.12.24740230 5.433512 1.2314180 0.9511901 0.4249051 #> ENSG00000001461.11.24742304 5.039812 1.3268172 1.2010861 2.1552969 #> ENSG00000001497.11.64754655 5.769157 0.7417616 0.7007832 2.3329780# Do some multidimensional scaling mds = cmdscale(dist(t(as.matrix(feat)))) plot(mds, type='n')text(mds, labels=rownames(mds))# limit to top 500 most variable "genes" sds = apply(feat, 1, sd) feat_500 = feat[order(sds, decreasing=TRUE)[1:500], ] heatmap(as.matrix(feat_500))