Getting started
## Loading required package: airway
library(SummarizedExperiment)
data(airway, package="airway")
airway
## class: RangedSummarizedExperiment
## dim: 64102 8
## metadata(1): ''
## assays(1): counts
## rownames(64102): ENSG00000000003 ENSG00000000005 ... LRG_98 LRG_99
## rowData names(0):
## colnames(8): SRR1039508 SRR1039509 ... SRR1039520 SRR1039521
## colData names(9): SampleName cell ... Sample BioSample
Exercises
- What is the
class() of airway? What does
the class tell you about the information stored about the features
(genes) in the dataset?
class(airway)
- How many features are in the data?
airway # it tells you
# but to get the result as a number:
nrow(airway)
- How many samples are present?
airway # it tells you
# but to get the result as a number:
ncol(airway)
- The assay information in the airway object contains the RNA-seq
counts. Access the count data for the 56th gene.
# check the names of the available assays
assayNames(airway)
# Both below are identical
assay(airway, "counts")[56, ]
assays(airway)[[1]][56, ]
- What are the
rownames() of airway? What do these
represent? What gene is represented by the first row of data?
head(rownames(airway))
# These are Ensembl gene identifiers
# ENSG00000000003 is the identifier of the "TSPAN6" gene
- What information is available about the samples?
colData(airway)
- How many samples were treated with dex? Untreated?
colData(airway) # and count
colData(airway)$dex # and count
table(colData(airway)$dex)
- How many different cell types were used?
table(colData(airway)$cell)
length(unique(colData(airway)$cell))
- What does
rowRanges(airway) give you? What is the
length of the object? What does each element of
rowRanges(airway) contain?
rowRanges(airway)
length(rowRanges(airway))
rowRanges(airway)[[1]]
- From the PubMed abstract,
we can see that the authors suggest that the DUSP1 gene is upregulated
by Dexamethasone treatment. The GeneCards
website tells us that the Ensembl gene id for DUSP1 is
‘ENSG00000120129’. Get the count data for this gene.
# get the assay data for DUSP1
dat = assay(airway, "counts")["ENSG00000120129", ]
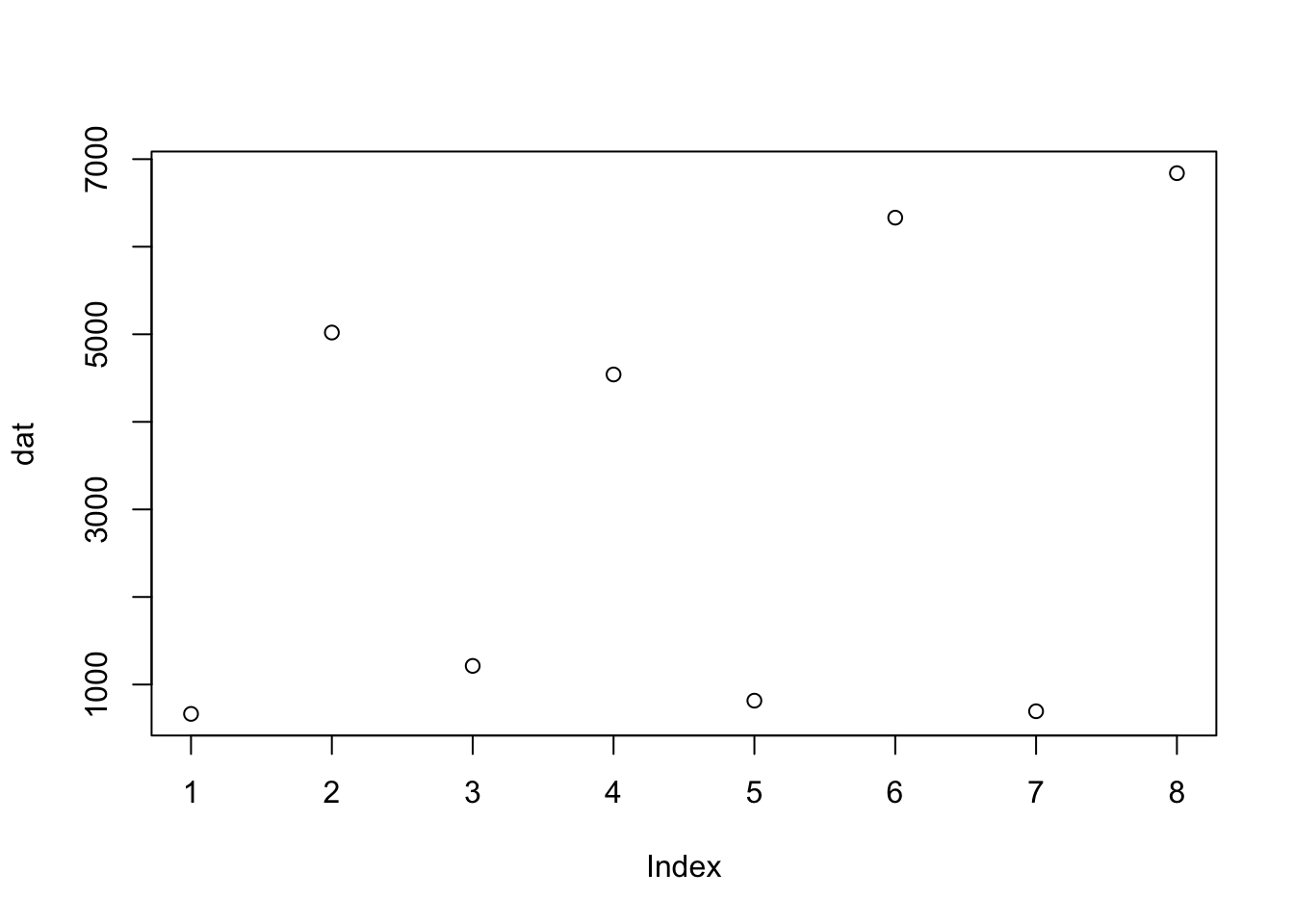
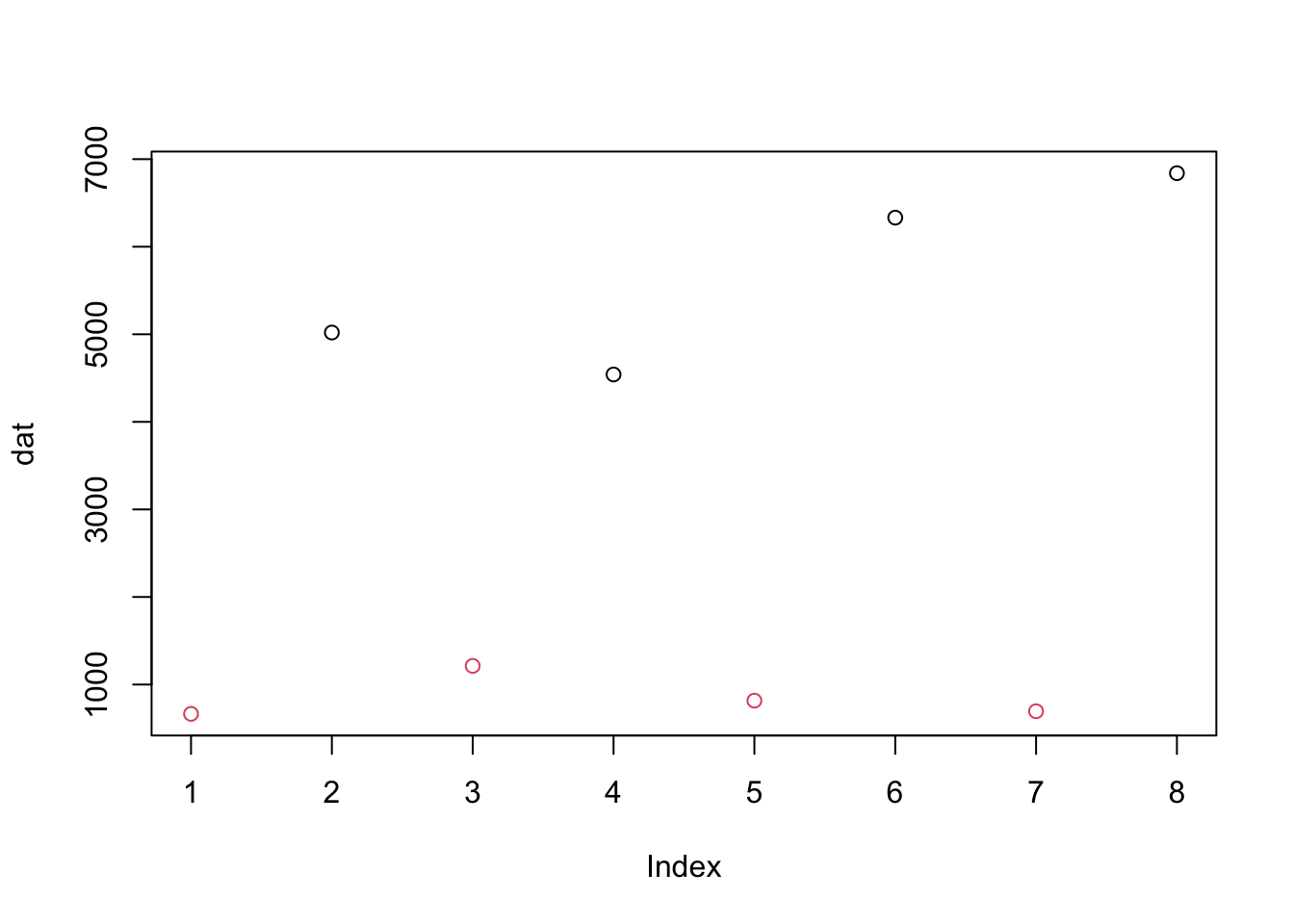
- Make a plot of the gene counts in the previous exercise. Color the
points based on the dexamethasone treatment.
# get the assay data for DUSP1
plot(dat)
dextrt = colData(airway)$dex
plot(dat, col=dextrt)
- Bonus: Load the logratios, gene and ORF information, and
sample information from the “DeRisi” data into a
SummarizedExperiment object. You will need to construct:
- A matrix of logratios as assay data.
- A data.frame of matching size and matching rows that contains the
symbol and ORF idendifiers.
- A data.frame with 7 rows, corresponding to the 7 columns of the
logratios.
sessionInfo()
sessionInfo()
## R version 4.2.0 (2022-04-22)
## Platform: x86_64-apple-darwin17.0 (64-bit)
## Running under: macOS Big Sur/Monterey 10.16
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.2/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats4 stats graphics grDevices utils datasets methods
## [8] base
##
## other attached packages:
## [1] airway_1.16.0 SummarizedExperiment_1.26.1
## [3] Biobase_2.56.0 GenomicRanges_1.48.0
## [5] GenomeInfoDb_1.32.0 IRanges_2.30.0
## [7] S4Vectors_0.34.0 BiocGenerics_0.42.0
## [9] MatrixGenerics_1.8.0 matrixStats_0.62.0
##
## loaded via a namespace (and not attached):
## [1] highr_0.9 bslib_0.3.1 compiler_4.2.0
## [4] jquerylib_0.1.4 XVector_0.36.0 bitops_1.0-7
## [7] tools_4.2.0 zlibbioc_1.42.0 digest_0.6.29
## [10] jsonlite_1.8.0 evaluate_0.15 lattice_0.20-45
## [13] rlang_1.0.2 Matrix_1.4-1 DelayedArray_0.22.0
## [16] cli_3.3.0 rstudioapi_0.13 yaml_2.3.5
## [19] xfun_0.30 fastmap_1.1.0 GenomeInfoDbData_1.2.8
## [22] stringr_1.4.0 knitr_1.39 sass_0.4.1
## [25] grid_4.2.0 R6_2.5.1 rmarkdown_2.14
## [28] magrittr_2.0.3 htmltools_0.5.2 stringi_1.7.6
## [31] RCurl_1.98-1.6

